New Population Genetic Distance Explorer
I’m thrilled to unveil the Population Genetic Distance Explorer, a new interactive tool to dive into the genetic diversity of human populations. Discover fascinating insights into migration patterns, population history, and genetic variation. Want to jump in? Scroll to the Get Started Today section to access the app link. To unlock the full post and explore the tool’s premium features, upgrade to a paid subscription below the paywall.
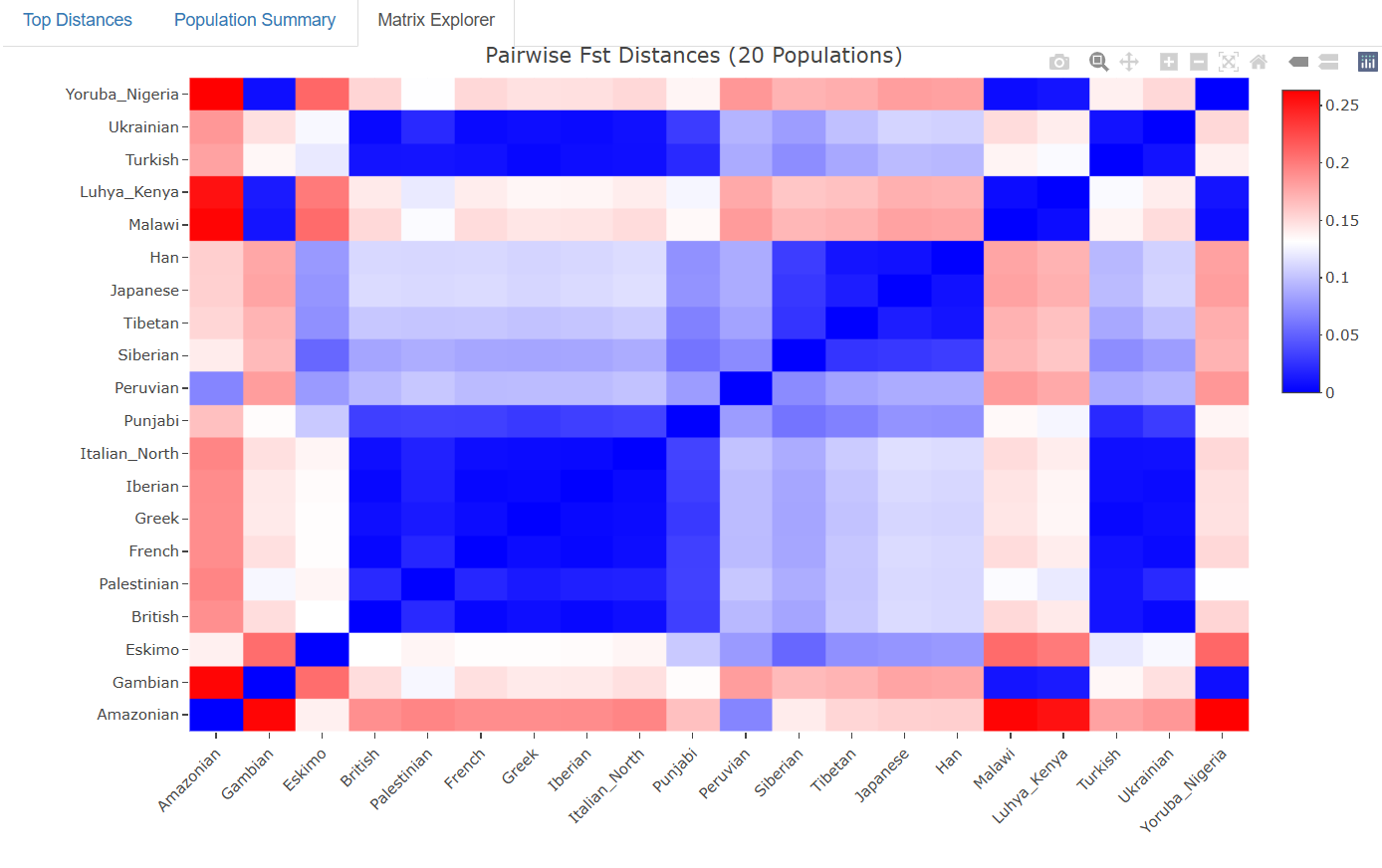
The visually oriented can use the Matrix Explorer, an interactive tool to visualize Fst distances between all possible pairs.
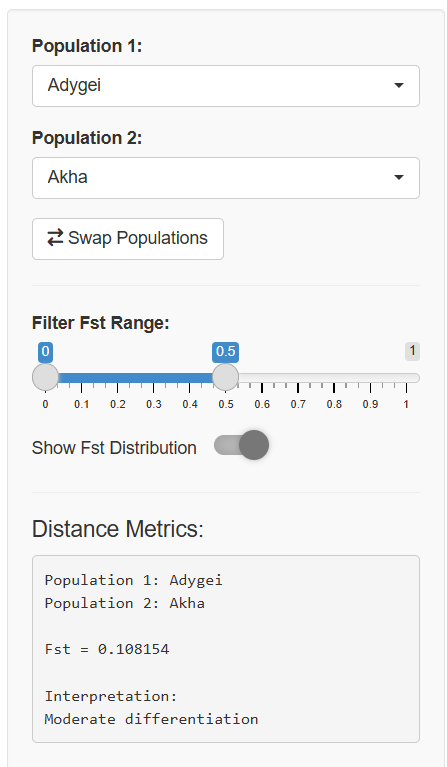
The core interactive component of the app is the Population Pair Selector, located in the sidebar panel.
Key Components of the Population Pair Selector:
Population 1 Dropdown: A searchable dropdown menu where you select the first population
Population 2 Dropdown: A searchable dropdown menu where you select the second population
Swap Button: An exchange icon that quickly swaps your two selected populations
Distance Metrics Display: Shows the Fst value and interpretation for your selected pair
Random Pair Buttons:
"Random High Fst Pair" for discovering populations with high genetic differentiation
"Random Low Fst Pair" for finding closely related populations
How it Works:
Select any population from the "Population 1" dropdown
Select any population from the "Population 2" dropdown
The app instantly calculates and displays:
The Fst value (genetic distance) between the two populations
An interpretation of what that value means biologically
Where this pair falls in the overall distribution of genetic distances
About the Data
The previously launched app used a dataset that I had personally curated. This new app utilizes data from the Allen Ancient DNA Resource (AADR), a comprehensive curated compendium of ancient human genomes compiled by Mallick and Reich (2023). For this application, I have focused exclusively on modern populations to provide a clear view of contemporary human genetic diversity.
The AADR represents one of the most extensive collections of human genomic data available, making it an ideal foundation for exploring population genetic structure. By leveraging this resource, I've created an intuitive interface that makes complex genetic data accessible to researchers, students, and genetics enthusiasts alike.
Two Versions for Different Needs
I understand that different users have different needs, which is why I am offering two versions of the application:
Free Version (20 Populations)
The free version provides access to a carefully selected set of 20 populations that represent major global populations:
Amazonian
Gambian
Eskimo
British
Palestinian
French
Greek
Iberian
Italian_North
Punjabi
Peruvian
Siberian
Tibetan
Japanese
Han
Malawi
Luhya
Turkish
Ukrainian
Yoruba
This diverse selection allows users to explore genetic distances across different continents and population groups, providing a solid introduction to population genetics.
Premium Version (115 Populations)
For paid subscribers, I offer access to an expanded dataset containing 115 populations, including:
Adygei
AfricanAmerican
AfricanCaribbean
Akha
Altaian
Amazonian
Armenian
Balochi
Bantu
Bashkir
Bedouin
Bengali
Biaka
Brahui
British
Burusho
Buryat
Caucasian
Central_Asian_Turkic
Central_Siberian
ChineseDai
Chukchi
Colombian
Dagestani
Dai
Dong
Druze
Esan
Eskimo
Faza_Bajun
Finnish
French
Gambian
Georgian
Greek
GujaratiIndian
Han
HanChinese_North
HanChinese_South
Hazara
Hungarian
Iberian
Indonesian
Iranian
Iran_Zoroastrian
Italian_Central
Italian_North
Italian_South
Japanese
Jewish
Kalash
Karitiana
Kazakh
Kenyan_Coast
KinhVietnamese
Kyrgyz
Luhya
Makrani
Malawi
Mandenka
Mayan
Mbuti
Mende
Mesoamerican
Mexican
Miao
Mongol
Mordovian
Mozabite
Nasioi
Naxi
Nepali
Nganasan
Nogai
Northwest_Caucasian
Orcadian
Oroqen
Other
Palestinian
Papuan
PapuaNewGuinea
Pathan
Peruvian
Pima
PuertoRican
Punjabi
Qiang
Russian
Sardinian
She
Siberian
Sindhi_Pakistan
Southeast_Asian
South_Slav
SriLankanTamil
Tajik
Tatar
TeluguIndian
Tibetan
Tu
Tubalar
Tujia
Turkish
Tuvinian
Ukrainian
Ulchi
US_Whites_Utah
Uyghur
Yakut
Yemeni
Yemeni_Highlands
Yemeni_Northwest
Yi
Yoruba
This expanded dataset offers a more comprehensive view of human genetic diversity, including many populations not covered in my previous applications. While there is some overlap with the population set used in the other recent app, this collection is partly distinct and offers new opportunities for exploration.
Key Features of the App
This Population Genetic Distance Explorer includes several powerful features:
Interactive Heatmap: Visualize genetic distances between populations in an intuitive color-coded matrix
Population Comparison: Select any two populations to see their specific genetic distance (Fst value)
Fst Distribution: Explore the distribution of genetic distances across all population pairs
Population Statistics: View summary statistics for each population
Filtering Options: Focus on specific ranges of genetic distances
Random Selection: Discover interesting population pairs with the random selection tools
Get Started Today
The free version of my Population Genetic Distance Explorer is available now. Simply visit the app page to begin exploring genetic distances between 20 selected populations.
For researchers, students, and genetics enthusiasts who need access to the full dataset with 115 populations, consider subscribing to the premium service. Paid subscribers gain access to the complete dataset along with additional features and updates.
The link to the full version with 115 populations is available below to paid subscribers.
Love my newsletter? Share it with friends and unlock exclusive rewards at 3, 5, or 25 referrals!